Challenges in Cancer Testing
Oncology molecular testing is challenging enough without the added complexity of difficult bioinformatics, variants of unknown significance, high cost, sample requirements and long turnaround times.
The MassARRAY® technology combined with the UltraSEEK™ and iPLEX® HS chemistries eliminate these challenges while maintaining the ability to target all the clinically relevant markers from liquid and tissue biopsies.
Tumor Tissue
iPLEX HS enables the detection of variants as low as 1% MAF from heterogenous solid tumor samples including FFPE tissue, core needle biopsies, FNA, cytology smears and fine needle aspirates.
Liquid Biopsy
UltraSEEK enables the detection of variants as low as 0,1% MAF from circulating cell free and tumor DNA (ctDNA) as well as from circulating tumor cells (CTC).
UltraSEEK™ EGFR Panel
- Enables the study of disease progression and resistance from CTCs and ctDNA across 6 clinically relevant variants in EGFR(see below)
- Detected at as low as 0.1% MAF
- Requires only 10 ng of DNA input
- from DNA to data in a single workday
- Test 96 samples in a single run

UltraSEEK™ Lung Panel
- Enables study of disease progression and resistance from CTCs and ctDNA
- Detecting 67 variants from a single blood draw at as low as 0.1% MAF in 5 genes (see below)
- Requires only 10 ng of DNA input
- from DNA to data in a single workday
- Test 8 samples in a single run

UltraSEEK™ Colon Panel
- Enables study of disease progression and resistance from CTCs and ctDNA
- Detecting over 100 variants from a single blood draw at as low as 0.1% MAF in 5 genes (see below)
- Requires only 10 ng of DNA input
- from DNA to data in a single workday
- Test 8 samples in a single run

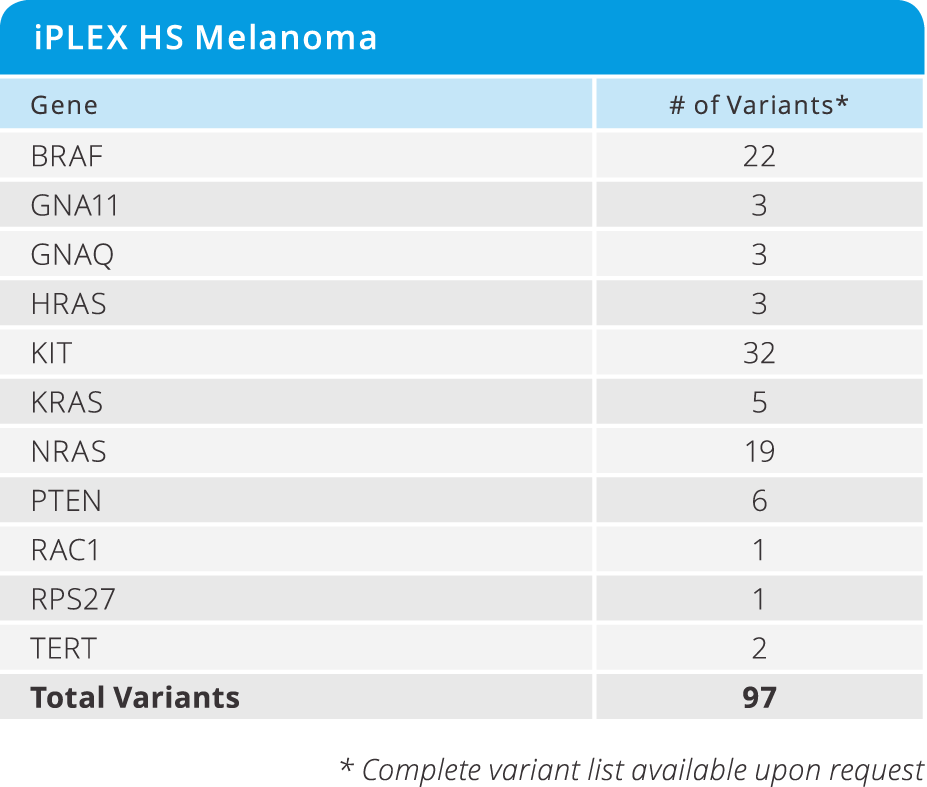
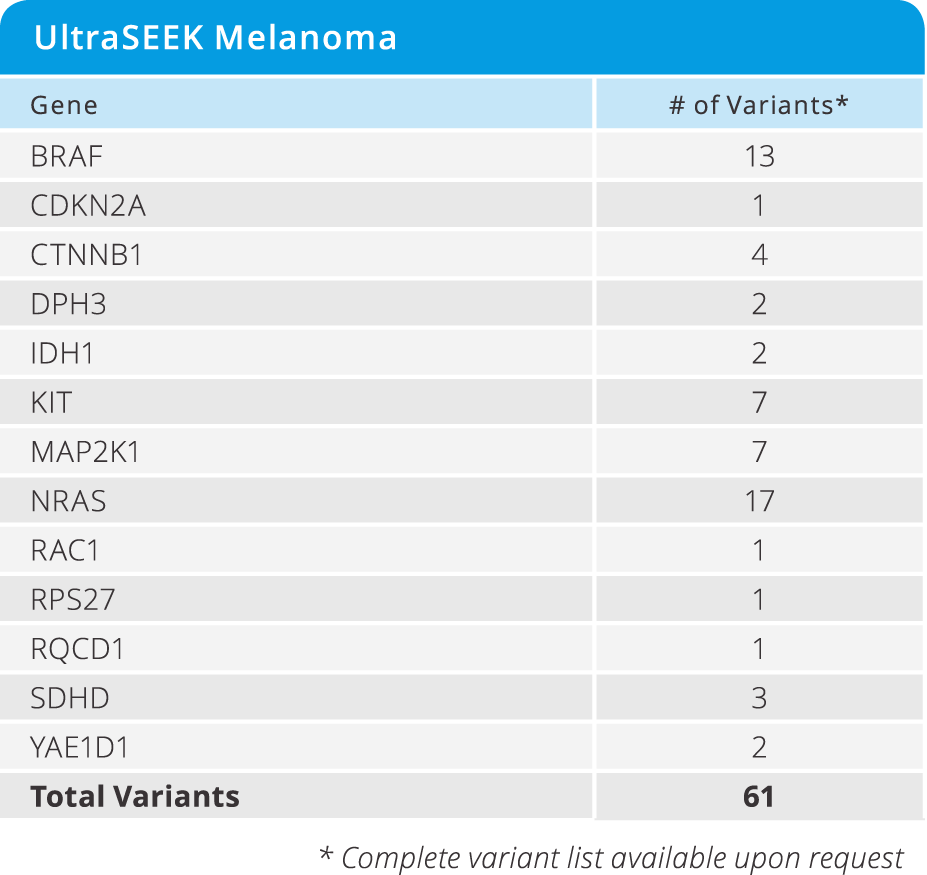
UltraSEEK™ Melanoma Panel
- Enables the study of disease progression and resistance from CTCs and ctDNA across 61 clinically relevant variants in 13 genes (see below)
- Detected at as low as 0.1% MAF
- Requires only 10 ng of DNA input
- from DNA to data in a single workday
- Test 8 samples in a single run
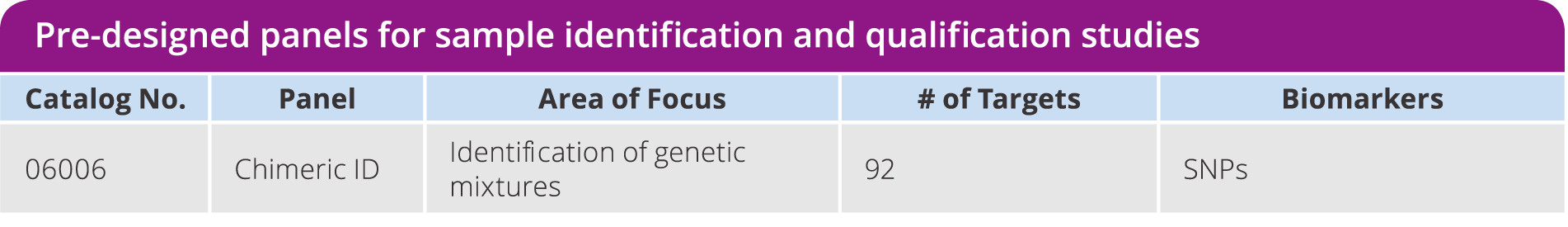
Chimeric ID Panel
Detect genetic mixtures, sample contamination, distinguish between maternal and fetal DNA, or monitor transplant success using SNPs instead of labor intensive short tandem repeat (STR) analysis. Utilize a pre-verified assay consisting of 92 SNPs, selected for high MAF across six major HapMap populations and not in linkage disequilibrium, which enables efficient identification of sample mixtures as low as 2%. Validated against the current gold standard STR method.
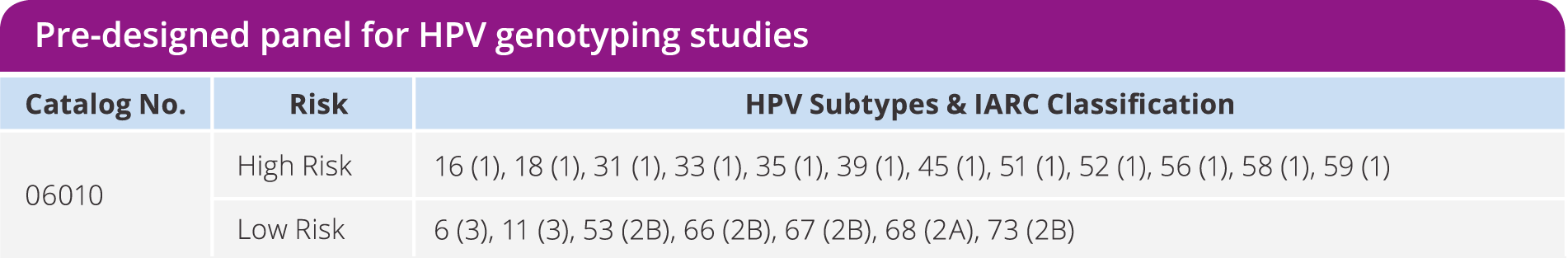
HPV typing
Enhance research in head and neck, gynecological, and dermatological cancers by identifying 19 different HPV subtypes. Utilize minimal input DNA to screen samples with limited availability and perform confirmatory testing.
All assays are for research use only, not for use in diagnostic procedures.
Need a unique solution? SEQ-IT can help with custom assay design service.
Contact: massarray@seq-it.de